January 28, 2026
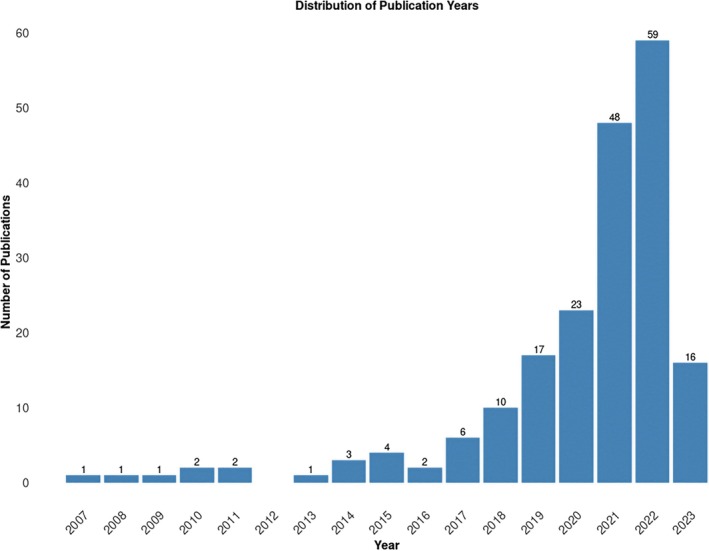
Machine learning (ML) offers opportunities to overcome limitations of conventional survival analyses, which are commonly found in cancer studies. It becomes unclear whether they consistently outperform traditional statistical methods and whether one particular ML strategy may outperform others for survival analysis. The present study aimed to systematically review the literature around this emerging topic. The review included 196 studies, from which 39 comparable studies were used for the analysis. The authors focused as they said on rigorous selection criteria and more specifically on time‐to‐event outcomes, so that their review provides insight into the comparative performance of machine learning, deep learning and classical statistical approaches.
The ML techniques are useful for analyzing high dimensional data that are often encountered in survival analysis. They can capture complex, non‐linear relationships between covariates and survival outcomes, unlike traditional statistical models. Several strategies have already been adapted for survival analysis and were found throughout this systematic review. These include regularization of the Cox proportional hazards (CPH) model, approaches based on survival trees, boosting algorithms, support vector machines, shallow neural networks and deep learning. The regularized methods added to the CPH add a penalty term to the likelihood to allow for high-dimensional data. Survival trees, like random survival forests where a subset of features is randomly selected at a node before the next split. Boosting is another ML method which combines predictions from multiple base learners. For survival analysis, it uses two main methods: model-based boosting, where residuals are computed and learners are fit to the residuals, and likelihood-based boosting, where weak learners are fitted by the negative gradient of the log-likelihood with respect to the parameters of the current model. Boosting in survival analysis can follow the traditional tree‐based structure similar to the original boosting methods, or it can be based on another modelling structure, such as the CPH model. Hyperparameters for such models include the learning rate parameter (ν) and either the number of iterations or the stopping criterion. Support vector machines (SVM) for classification involves creation of hyperplane decision boundaries to separate data into classes. There are two ways of survival SVM, the regression approach and the ranking approach. Kernel functions can be used to capture the non-linear relationships between covariates and survival time. Neural networks (NN) for survival analysis use artificial neural network architectures to predict time-to-event outcomes and also model complex, non-linear relationships in data. Deep learning is a subset of ML that is focused on NN but has a large number of deep architectures. One popular one, DeepSurv combines CPH regression with neural networks.
Per the results of their systematic analysis, they had found that Approximately 55% of the studies compared ML models to a traditional statistical methodology or a current state‐of‐the‐art model (SoA). The most prevalent ML modelling method was random survival forest (RSF), which was considered in more than 55% of the studies and as the only ML model in 19% of studies. Fourteen studies also considered other tree‐based methods, such as single survival trees and conditional survival forests (CSF). Neural network‐based methods were the next most prevalent, most of which being deep learning structures (approximately 40% of all the studies), while shallow architectures were considered in 15% of studies. Evaluation metrics were considered under 3 main headings: discrimination (Harrell’s c-index and Brier score were used), calibration (ROC or calibration metrics), and predictive performance.
Overall, they summarized outcomes from 196 studies relating to use of ML for survival analysis in cancer. The authors also concluded that deep learning and multi‐task methods performed better than classical survival analysis methods, like CPH. Their review yielded two major findings. One was that improved predictive performance was obtained from ML methods across all cancer types, with the exception of cervical cancer. The other was in the heterogeneity of ML models used, and variability in the choice of ML pipelines. A majority of papers reported on output for a random survival forest or other tree‐based method (58%, 114/196 studies), deep learning frameworks (42%, 83/196 studies) or regularized Cox models (36%, 71/196 studies). Most studies did not consistently compare the ML methods to conventional survival models nor always make comparisons between several ML models, limiting the number of studies to draw conclusions from. Direct comparisons between ML models were also limited by how they reported outcomes of interest as well as performance metrics used.
Written by,
Usha Govindarajulu
Keywords: machine learning, survival analysis, deep learning, Cox proportional hazards regression, random survival forests, neural networks, prediction, calibration, discrimination,
O’Donnell A, Cronin M, Moghaddam S, Wolsztynski E. A Systematic Review on Machine Learning Techniques for Survival Analysis in Cancer. Cancer Med. 2025 Nov;14(22):e71375. doi: 10.1002/cam4.71375. PMID: 41264402; PMCID: PMC12633653.
https://pmc.ncbi.nlm.nih.gov/articles/PMC12633653/
https://cdn.ncbi.nlm.nih.gov/pmc/blobs/c3c1/12633653/4cbaa88359da/CAM4-14-e71375-g003.jpg