June 18, 2025
The goal of this paper was to bridge gaps in understanding how to use machine learning (ML) methods for survival by presenting a comprehensive study comparing various ML methods for dynamic survival analysis. They sought to provide researchers and practitioners in the field of cognitive health, and healthcare in general, with a clearer understanding of the strengths and weaknesses of these methods and the nuances of their implementation. They said their main contributions of this study were (i) the comparison of methods for dynamic survival analysis across diverse scenarios, including both statistical approaches, such as MFPCA, and neural network-based methods, (ii) the separate investigation of the contributions to the system performance of longitudinal and survival models by testing multiple combinations and (iii) the comparison of several landmarking strategies that can be used during model training.
For dynamic prediction, they discussed using landmarking but building separate survival models at each landmark time. Joint modeling of longitudinal and survival data also exist. They then came up with a two-stage modeling approach, by which they can effectively combine different longitudinal and survival models. In the first stage, a longitudinal model is used and in the second stage, predictions of the longitudinal model are incorporated into the survival model. They felt this strategy works well to avoid joint modeling complexities.
They discussed several landmark approaches: none, strict, super, and random. None uses all data to make decisions. The strict uses a separate model trained at each landmark time but risks losing data is not used in each landmarked dataset. The super also does what the strict does but ends up combining each separate landmarked dataset into one large dataset. Random would at each iteration randomly sample a subject and temporarily truncate the longitudinal data for this subject. It reduces computational burdens.
Longitudinal models were discussed. The multivariate functional principal components analysis (MFPCA) is a method to capture temporal dynamics in longitudinal data by representing them in a lower dimensional space using functional principal components analysis but it does not handle the survival data. Recurrent neural networks (RNN)s are a class of neural networks suited for sequential data analysis and then can be applied to longitudinal data. Survival models that were discussed were Cox proportional hazards regression for which the covariate is the ith subject from the longitudinal model, the random survival forest (RSF), and neural network.
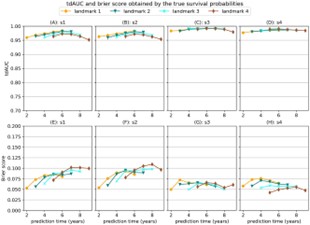
For evaluation, they setup 4 simulation scenarios and one real dataset application. To assess model performance, they used an inverse probability of censoring weighting (IPCW) time-dependent area under the curve, an IPCW Brier Score which used a mean squared error calculation as the difference between observed and predicted risk, which is calculated as 1 minus the ratio of predicted survival for the ith outcome to the predicted survival for the landmarked ith outcome.
The simulations showed the Cox model fit baseline data well but after an interaction term was added or different slopes were added for different subjects, then the RSF or RNN performed better. The methods performed the worst when there was no landmarking and better when there was landmarking. The results on the real dataset had less conclusive results. The strict or no landmarking resulted in worse performance amongst methods. The RSF frequently produced strong results. The more complex methods in general did not fare well on the real dataset application. They in general found that the qualitative conclusions drawn from their experiments did not strongly depend on choice of fixed hyperparameters. Another consideration was computational efficiency, the RNNs benefited from GPU acceleration. The baseline and last visit benchmarks were faster than the other methods while the MFPCA was notably slower. Among landmarking strategies, the no landmarking and random landmarking methods were the most efficient.
In conclusion, some form of landmarking is often beneficial. The RNN model demonstrated strong performance across all synthetic datasets. For survival models, the RSF showed robustness and versatility. In general, they suggested that understanding the trade-offs amongst these choices was essential for successfully conducting dynamic survival analysis.
Written by,
Usha Govindarajulu
Keywords: dynamic survival, landmarking, Cox model, machine learning, RSF, RNN, MFPCA, IPCW
References:
De Swart WK, Loog M, and Krijthe JH (2025) “A comparative study of methods for dynamic survival analysis” Front. Neurol., Sec. Artificial Intelligence in Neurology, Volume 16 – 2025 | https://doi.org/10.3389/fneur.2025.1504535