March 15, 2023
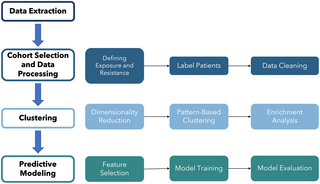
In a recent article that appeared in Science News, a teach from Johns Hopkins University discussed their proof of concept study to show how their machine learning system used electronic health data to make predictions to determine who would be most resistant to COVID-19, even after exposure to the Sars-CoV-2, the actual virus which causes this disease. The actual study results were published in PLOS ONE. The machine learning algorithm trained on data obtained from Johns Hopkins COVID-19 Precision Medicine Analytics Platform Registry, which has information on patients seen or suspected of having COVID-19 infection by those within the JHU health system. Information on demographics, medications, and comorbidities. They used training and test sets split on two groups, associated with resistance or not associated.
In the PLOS one article, much more detail is provided about methods used. The other used a standard logistic regression and machine learning methods, random forests and XGBoost, The “optimized” models were evaluated then on testing subsets from what they called the “low-confidence exposure cohort” and the HHI testing subset. They describe using a Shapley Additive exPlanations method to evaluate the relative importance of each feature. They did not describe how this method works. However, this seemed to become consequential in seeing how each feature impacted each outcome.
They found certain diagnostic code associated with the infection like ICD-10 codes associated with screening and parasitic disease but the only diagnosis they found associated with increased resistance to COVID-19 was personal history of malignant neoplasm. They also found greater number of comorbidities and medications in the non-resistant cohort compared to resistant persons. They discussed how they had obtained p-values through Monte Carlo simulation for the testing set of all models indicating the results were all statistically significant. These conclusion was not clear to understand. They also said their XGBoost model performed better than all the models even though its AUROCs were in the range of 0.6. Overall, they concluded their ML methods worked well to detect patients who will end being resistant to emerging infection.
In their limitations section, they discussed not knowing which patients were using masks or not throughout the study and also they were limited to patients who visited within the JHU medical system. Furthermore, patients labelled as resistant could have tested positive for COVID-19 outside of their time frame. These limitations they assessed do not seem minor at all and definitely could have skewed their results. Their results and their conclusions were not all together conclusive of their findings and a better controlled analysis was needed without so many biases and confounders. Unfortunately, not using statistical modelling to control for these have hampered these results.
Written by,
Usha Govindarajulu
Keywords: COVID-19, machine learning, logistic regression, XGBoost, random forests
References
Johns Hopkins Medicine. “Research team creates statistical model to predict COVID-19 resistance: Proof-of-concept study shows promise for machine-learning system that uses electronic health data to make its predictions.” ScienceDaily. ScienceDaily, 22 February 2023. <www.sciencedaily.com/releases/2023/02/230222210542.htm>.
Kai-Wen K. Yang, Chloé F. Paris, Kevin T. Gorman, Ilia Rattsev, Rebecca H. Yoo, Yijia Chen, Jacob M. Desman, Tony Y. Wei, Joseph L. Greenstein, Casey Overby Taylor, Stuart C. Ray. Factors associated with resistance to SARS-CoV-2 infection discovered using large-scale medical record data and machine learning. PLOS ONE, 2023; 18 (2): e0278466 DOI: 10.1371/journal.pone.0278466
https://journals.plos.org/plosone/article/figure/image?size=inline&id=10.1371/journal.pone.0278466.g001