February 1, 2023
In an article recently published in Biometics, Huang et al discuss how to adjust for publication bias in meta analyses via using inverse probability weighted adjusted measures of meta-analyses. The authors were motivated by the fact that meta-analyses are used commonly in the medical literature, but none of the commonly used techniques to date adjust for publication bias, which has been rampant in the field. The authors focused their efforts on clinical trial registries.
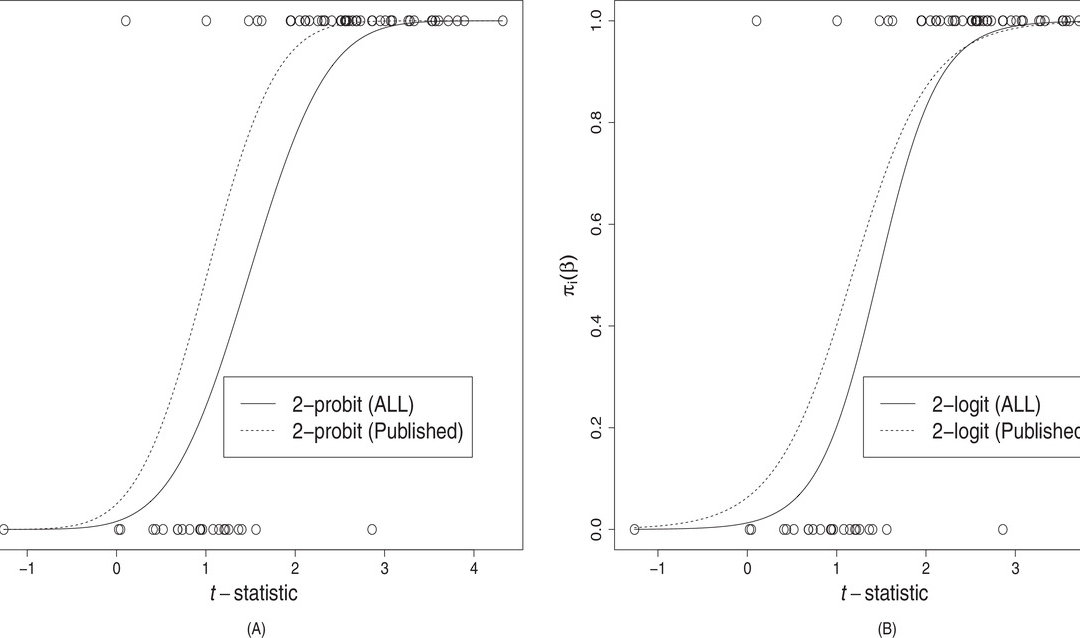
The authors developed an estimating equation and treated the publication bias like a missing at random problem. They first developed a selection model, generally based on a one or two parameter logistic function using a t-type statistic as the main predictor, where t is defined as a ratio of y (log hazard of treatment effect) and σ2 (variance of treatment effect) from a particular study. They then created an inverse probability weighted estimator which is similar to that of the propensity score for missing data or causal inference, where they incorporated a binary indicator for publication. The motivation for the author’s derivation of this weighted estimator is not well explained and seems to heavily rely on already developed theory from causal inference. They then show the parametric bootstrap to obtain confidence intervals around their estimator. Finally, regarding other measures of between study heterogeneity, the H2 and the I2 statistics, they came up with their own IPW version of these using an IPW version of the Q-statistic, which is then employed in the formulas for H2 and I2.
Their simulation studies were conducted both on the one-parameter and the two-parameter section functions. They randomly generated four datasets based on either the one or two-parameter estimating functions. They compared their method against the Copas sensitivity method and the standard mixed-effects model. They presented results for estimating µ for which they sound the Copas sensitivity analysis and their IPW method could reduce bias. For τ2, their IPW version of DerSimonian-Laird estimator had smaller bias than the τ2DL estimator. Then they show results for their estimator. They said if there was severe selective publication bias then their method did not do well but it had smaller biases in most of their scenarios.
The authors concluded with that their method is more simplistic and easier to employ than previous methods of the same type. They also found their parametric bootstrapping for confidence intervals was sufficient. They again brought up the missing data issue and how the publication bias issue was long recognized as one, but as far as I can see in their paper, this issue was not well described in their methods section in terms of the development and/or it seems they definitely took from this methodology for propensity scores. They did admit with their method that it was hard to chose which selection method to use within it so probably a sensitivity analysis would have to be done by future users of their method. All in all, this was not a well laid out description of their proposed estimator.
Written by,
Usha Govindarajulu
Keywords: IPW, publication bias, meta-analyses, DerSimonian-Laird, propensity scores, missing data, Copas sensitivity
References
Huang, A., Morikawa, K., Friede, T., & Hattori, S. (2023) Adjusting for publication bias in meta-analysis via inverse probability weighting using clinical trial registries. Biometrics, 00, 1– 13. https://doi.org/10.1111/biom.13822
https://onlinelibrary.wiley.com/cms/asset/d614a47a-ec3d-430d-b3f5-a1cacc4ba1f8/biom13822-fig-0002-m.jpg