July 5, 2023
The restricted mean survival time (RMST) is a concept that has been invented in survival analysis as an alternative to the hazards ratio, which can be difficult to interpret and also, as derived from Cox model, can have difficulty fitting into the proportional hazards assumption. The RMST is defined as the mean survival time from followup to some specified time, τ (Uno, 2014). A problem that the authors note is that the RMST is that it is defined within a specific time window. Therefore, a conditional RMST (cRMST) was invented to handle dynamic changes within the RMST.
The authors defined the equation for the cRMST and also show how it can be computed at landmark time points within some time interval. In addition, they showed how a landmark dataset can be constructed to incorporate fixed and time-varying covariates in what they call a generalized linear model where the regression parameters are derived from a generalized estimating equation (GEE) method. From there they derived a dynamic RMST model where they can calculate the survival at multiple time points unlike the usual RMST where it is calculated up to a specific time point. They did this first by creating the landmark datasets per each landmark time point and stacking up the datasets into what they called a “super prediction dataset” where the sample size corresponds to each individual. Now again they fit a generalized linear model on this dataset, a linear model and employing basis functions on the parameter estimates which could be used for smoothing. Then to estimate the parameter estimates, they again employed a GEE, this time with the vector of basis functions. Within this framework, they used a working covariance matrix and also a sandwich estimator for the variance-covariance matrix. Their cRMSTd is defined as the difference between two groups.
In a Monte Carlo simulation study, they explored how accurate the estimation of the cRMSTd performed under both proportional hazard assumptions (PH) and non-PH assumptions. They generated survival time for N=1,000,000 subjects and calculated a true cRMST by averaging the RMST of the subjects who were still at risk at s. They then calculated the following indicators: bias, relative bias, root mean squared error (RMSE), relative standard error, and empirical coverage probability. They found the cRMSTd had small bias under all scenarios and the RMSE decreases with increasing sample size and decreasing censoring rate. Also, most of their relative standard error was approximately equal to 1 and most of the CP fell within a reasonable range, they did not state the CP that they found on average.
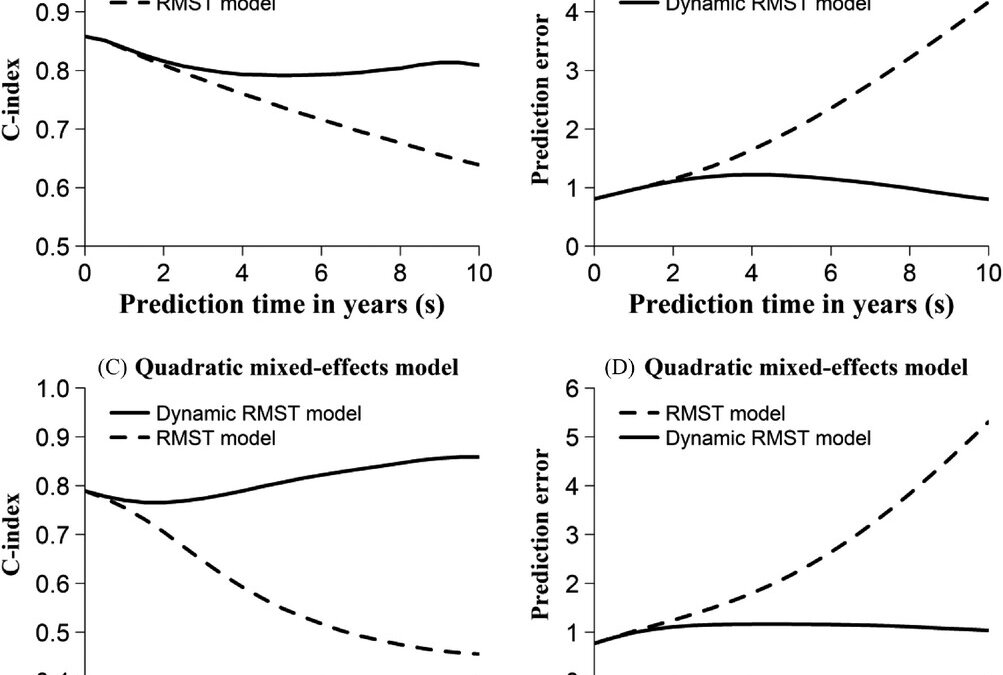
They further conducted simulation studies on the dynamic prediction model: to verify accuracy of regression coefficient estimation and to evaluate prediction performance of the model. The data for this was generated based on a joint model of longitudinal data and survival data. In the longitudinal biomarker data they included noisy measurement erors and employed linear mixed-effects model and/or quadratic mixed effects model with random intercept and slope or additonal time term for the quadratic time effect. The survival model was Weibull based and they gave no justification for their choice of the scale or shape parameters. They used a natural cubic spline basis function in this simulation. They found their regression coefficients were estimated with minimum bias and good coverage as well as low root mean squared errors. For the predictive performance, they employed Harrell’s C-index (Harrell, Lee and Mark, 1996) and the prediction error (Tian et al, 2007). They found the prediction performance of their dynamic RMST improved as the sample size increased as measured by the C-index greater than 0.76 and the the prediction error being relatively low.
Finally, in terms of a real data example, they used data from 407 patients with chronic kidney disease who receive renal transplantations in Belgium between 1983 and 2000. They employed their dynamic prediction on 5-years for the cRMST. However, they did employ a backward stepwise model-selection procedure on the quasi-likelihood under the independence model criterion. This seemed strange to use this very biased selection method after all the techniques they were employing. They then evaluated this data with their dynamic prediction model and explored the dynamic effects of covariates on graft life expectancy.
The authors have concluded that their method is robust and more useful than the RMST and they have also recommended an estimation method using pseudo-observations and the statistical inference concerning the difference between two cRMSTs. They also mentioned a different smoothing method besides natural cubic spline function could be used here and same also for the working correlation matrix.
Written by,
Usha Govindarajulu
Keywords: survival, RMST, hazards, cRMST,GEE, GLM, basis function, natural cubic spline
References
Harrell, F.E., Lee, K.L. & Mark, D.B. (1996) Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Statistics in Medicine, 15, 361– 387.
Tian, L., Cai, T., Goetghebeur, E. & Wei, L.J. (2007) Model evaluation based on the sampling distribution of estimated absolute prediction error. Biometrika, 94, 297– 311.
Ying Z, Zhang C, Hou Y, and Chen Z (2023). “Analysis of dynamic restricted mean survival time based on pseudo-observations” Biometrics. https://doi.org/10.1111/biom.13891.
https://onlinelibrary.wiley.com/cms/asset/b242cff4-9d94-4214-a907-ac601c1036fc/biom13891-fig-0001-m.jpg