August 28, 2024
The authors have proposed using imputed data from subdistribution weights to train on machine learning methods for competing risk data. The authors focused on the random survival forest (RSF) method which it stars with a number of bootstrap samples and grows a survival tree on each sample. Each tree would have then be split into noes where the best split is selected. The splitting rules per their application was done using the log-rank test and finally the tree stops growing when the minimum number of observations or no increase with respect to splitting criteria is even possible. A cumulative hazard function would have been calculated at each node of the tree. In order for the algorithm to use split rules they propose to first impute censoring times in cases competing events were observed. They then estimated subdistribution weights which were based on the censoring mechanism in the dataset.
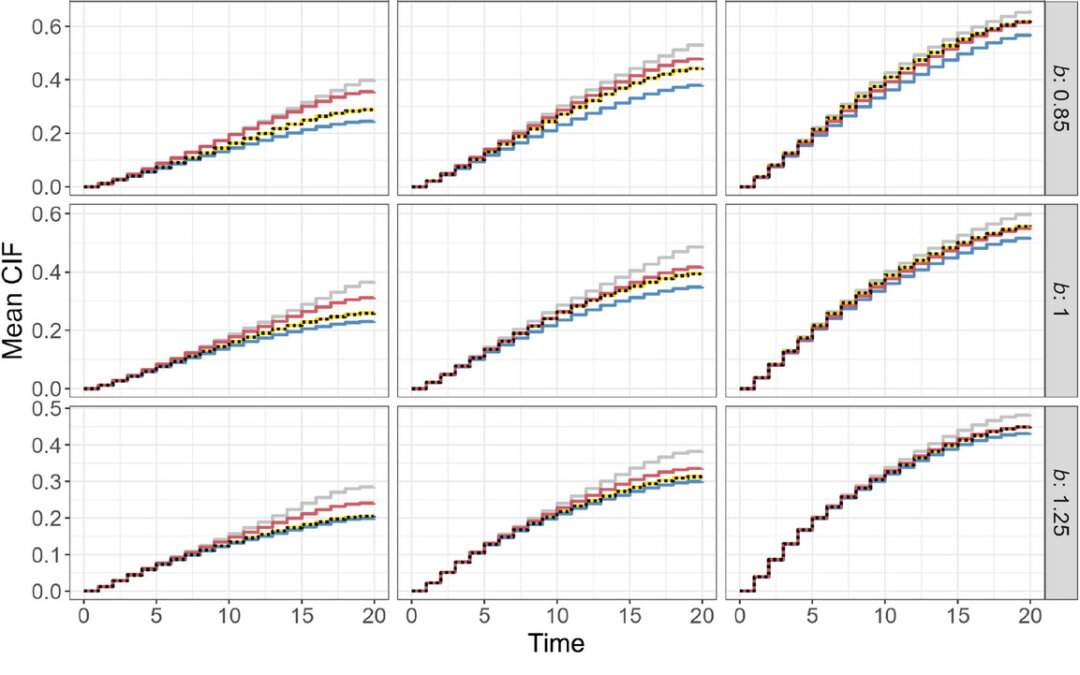
For their implementation, they used two R package, one to handle the imputation, ranger, and another to calculation a life table estimate of the censoring survival function, discSurv. They then implemented simulations to see if subdistribution-based imputation in case of competing events could improve the estimation of the cumulative incidence function (CIF) in RSF compared to ignoring competing risks. Within this they had to create a tree-like covariate-risk relationship. They used several measures to assess performance. They used a calibration graph to assess agreement between the reference and the estimated CIFs. They also used the c-index to assess discrimination of the different model fits on the test data. Finally, they used a Brier score to compare predictive performance of the different approaches of imputing directly in the nodes vs the methods that do not directly impute in the RSF.
They ran these approaches through simulations as well as on a real dataset, GCKD study data. The simulation study showed that the CIF was well established when the imputation takes place outside the random forest on training data. The other strategies produced an under or over-estimation of the CIF in the simulation study but performed better in the GCKD study which had larger sample sizes. Basically, their proposed single-imputation strategy allowed converting the competing-risk data into a single-event setting. RSF was then available for this setting.
Written by,
Usha Govindarajulu, PhD
Keywords: survival analysis, RSF, imputation, missing data, competing risks, CIF
References
Behnig C, Bigerl A, Wright MN, Sekula P, Berger M, and Schmid M (2024) “Random Survival Forests with Competing Events: A Subdistribution-Based Imputation Approach” Biometrical Journal, https://doi.org/10.1002/bimj.202400014 .
https://onlinelibrary.wiley.com/cms/asset/f2f8c7bb-2173-42db-ba7e-4e87b85955be/bimj2610-fig-0002-m.png